Cultivation of Magnetospirillum sp. XM-1, a New Strain of Magnetotactic BacteriaUpdate time:12 31, 2015
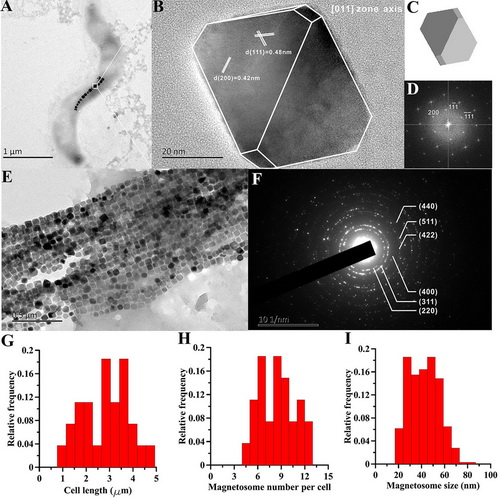
Magnetotactic bacteria (MTB) are a group of prokaryotes that can synthesize intracellular magnetite (Fe3O4) or/and greigite (Fe3S4) particles. These particles, called magnetosomes, are usually arranged in one or multiple chains within a cell and help MTB navigate along the magnetic field. Because the biotechnological applications of magnetosome magnetite have been widely considered in nano-material for magnetic separation, cancer detection, magnetic targeting hyperthermia, toxic metal bioremediation, and even in fields such as magnetic sensors and magnetic memory, cultivation and characterization of new MTB strains from nature environments are desirable. Pure culturing of MTB is necessary for understanding their physiology, evolution and biomineralization. Efforts to cultivate MTB suggest that magnetosomes synthesized by different MTB strains are diverse; thus they may have specific utilization in various domains. Magnetospirillum within the Alphaproteobacteria are considered model organisms for magnetite magnetosome biomineralization. Recently, PhD candiadate Yinzhao Wang and his supervisor Prof. Yongxin Pan from the CAS Key Lab of Earth and Planetary Physics, IGGCAS and coworkers successfully isolated and cultivated one novel Magnetospirillum sp. strain XM-1. Transmission electron microscope (TEM) observations show that XM-1 synthesizes a single intact chain with an average of 10 magnetosomes per cell with mean size and shape factor of 43.7 nm and 0.85, respectively (Fig. 1H and I). High-resolution TEM (HRTEM) observations reveal that magnetosomes are truncated, octahedral magnetite (i.e., slightly elongated cuboctahedrons) (Fig. 1B to E). Wang and coworkers performed a wide range of magnetic measurements to characterize the magnetosome chains (Fig. 2) and demonstrated that they behave like non-interaction single domain particles, which is typical for most MTB. They also combined response surface methodology to define the optimal growth medium composition and the best productive conditions at certain prerequisite with rock magnetic measurements. The optimal concentrations for the three factors derived from the model are 17.32 mM for NaNO3, 20.18 mM for sodium succinate and 2.13 mM for ferric citrate, which is template named as XM-C medium. Surprisingly, in a pH static fermenter, the cell growth increased to a maximum point (0.37 g/L cell dry weight, 7.1 mg/L magnetosome dry weight) within 48 h growth. This work has now been published in FEMS Microbiology Letters.
Figure 1. Transmission electron microscopy analyses of the XM-1. (A) typical TEM micrograph of strain XM-1; (B) magnetosome imaged by HR-TEM (crystal structure is shown by white line along the magnetosome); (C) calculated 3D model viewed from 011 axis; (D) FFT pattern of the crystal; (E) and (F) the TEM picture and SEAD pattern of the extracted magnetosome, respectively. Plots (G), (H) and (I) show the histograms of cell length, magnetosome number per cell and magnetosome size of strain XM-1, respectively.
Figure 2. Magnetic properties of XM-1 cell samples. Ferromagnetic resonance (A), low temperature magnetic measurements (B), room-temperature hysteresis loop (C), Wohlfarth-Cisowski test (D) and FORC diagram (E).
|
Contact
PAN Yongxin
Division: Earth's Deep Structure and Process Goup: Geomagnetic field and deep Earth interior processes Phone: 86-10-82998406 E-mail: yxpan@mail.iggcas.ac.cn Reference
|
-
SIMSSecondary Ion Mass Spectrometer Laboratory
-
MC-ICPMSMultiple-collector ICPMS Laboratory
-
EM & TEMElectron Microprobe and Transmission Electron Microscope Laboratory
-
SISolid Isotope Laboratory
-
StIStable Isotope Laboratory
-
RMPARock-Mineral Preparation and Analysis
-
AAH40Ar/39Ar & (U-Th)/He Laboratory
-
EMLElectron Microscopy Laboratory
-
USCLUranium Series Chronology Laboratory
-
SASeismic Array Laboratory
-
SEELaboratory of Space Environment Exploration Laboratory
-
PGPaleomagnetism and Geochronology Laboratory
-
BioMNSFrance-China Bio-mineralization and Nano-structure Laboratory

 Print
Print Close
Close