Metagenomic analysis reveals unexpected subgenomic diversity of magnetotactic bacteria within the Phylum NitrospiraeUpdate time:02 25, 2011
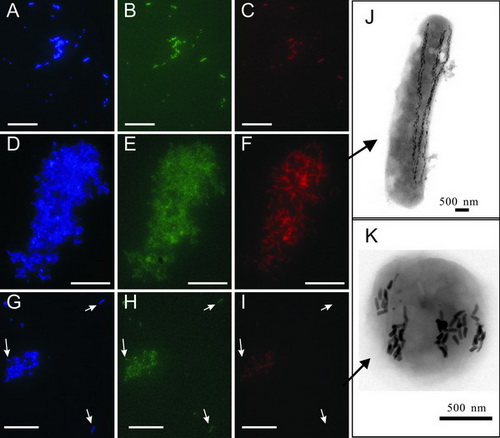
Fluorescence in situ hybridization (A to I) and transmission electron micrographs (J and K) of “Ca. Magnetobacterium bavaricum”-like Postdoctor LIN Wei and professor PANG Yongxin apply a targeted metagenomic approach to investigate magnetotactic bacteria (MTB) within the phylum Nitrospirae in Lake Miyun near Beijing, China. Five fosmids containing rRNA operons are identified. Comparative sequence analysis of a total of 172 kb provide new insights into their genome organization and revealed unexpected subgenomic diversity of uncultivated MTB in the phylum Nitrospirae. In addition, affiliation of two novel MTB with the phylum Nitrospirae is verified by fluorescence in situ hybridization. One of them is morphologically similar to “Candidatus Magnetobacterium bavaricum,” but the other differ substantially in cell shape and magnetosome organization from all previously described “Ca. Magnetobacterium bavaricum”-like bacteria. The achievement is published on Applied and Environmental Microbiology. Lin et al. Metagenomic analysis reveals unexpected subgenomic diversity of magnetotactic bacteria within the Phylum Nitrospirae. Applied and Environmental Microbiology, 2011, 77(1): 323-326
|
Contact
Related Articles
Reference
|
-
SIMSSecondary Ion Mass Spectrometer Laboratory
-
MC-ICPMSMultiple-collector ICPMS Laboratory
-
EM & TEMElectron Microprobe and Transmission Electron Microscope Laboratory
-
SISolid Isotope Laboratory
-
StIStable Isotope Laboratory
-
RMPARock-Mineral Preparation and Analysis
-
AAH40Ar/39Ar & (U-Th)/He Laboratory
-
EMLElectron Microscopy Laboratory
-
USCLUranium Series Chronology Laboratory
-
SASeismic Array Laboratory
-
SEELaboratory of Space Environment Exploration Laboratory
-
PGPaleomagnetism and Geochronology Laboratory
-
BioMNSFrance-China Bio-mineralization and Nano-structure Laboratory

 Print
Print Close
Close